Clinical challenges and assesments :
- Framework for Neurosphere Modeling under Phase-Contrast Microscope

- Integrated Virtual Cognitive Microscopy and Microscopy Mining for Breast cancer Grading support in hystopathology

- Cognitive Explolration of histopathological images and Visual Reasoning

- Parkinson's disease prognosis using MRI multimodalities fusion

ICV4NSC project & TRACKING DEMO & IMAGE INFORMATICS of Stem Cells
One of the major recent research field in the biomedical domain lies in the understanding of stem cells. Sptem cells are characterised and identified by the expression of three properties: (i) the capacity to renew their population over a large period of time, (ii) the capacity to produce specialised cells of a particular tissue, and (iii) participate into the regeneration of tissue after injury. Present in high concentration during the foetal phase as they participate to its development, their number drastically diminish once the individual reach the adult state. They remaining stem cells, renamed somatic (or adult) stem cells, are present in different tissue of the body, and their existence and role raised multiple interrogations: Where do they come from? Do they participate in the regeneration process of some tissue? Can they produce specialised cells of a different host tissue? Is there a link between somatic stem cells and cancer cells? In the case of neural stem cells, somatic stem cells of the central nervous system, their characterisation and culture, in vitro, is done through the Neurosphere Formation Assay (NFA) experiment proposed by Reynolds and Weiss in 1992. The experiment consists in isolating neural cells extracted from the specific tissue of the central nervous system and place them in suspension in a dish for a defined period of time (˜5 days). In the case of the isolated cells are generic, they will inevitably die after a certain time. If the isolated cells are neural stem cells, the expression of their renewal and differentiation properties will lead to the formation of spherical agglomerate structure of cells called neurosphere. The question we ask ourselves is: Could the observation and the understanding of the formation process of neurosphere bring information to understand the role of neural stem cells and help us to better control their culture in vitro ?
Leveraging our experience with traditional approach to biomedical imaging which rely on high content image analysis, this study proposes a method to determine the 3D cell configuration of a structure such as neurosphere, from a phase-contrast microscope 2D observation. Focussing on the structural aspect of the neurosphere, our work gathered methods from different field, from image processing and analysis, biomedical imaging, bioinformatics and artificial intelligence. Two modeling approaches, respectively using Delaunay mesh structure and evolution algorithm, are proposed to extract the cells configuration present in the microscope observation. Finally, the global framework allows the enhancement of neurosphere proliferation monitoring, an augmented reality support visualisation process and a tool for the extract of struc- tural information from the different neurosphere observation. It opens new perspective for neural stem cells research, in term of observation and high-content analysis.
Stephane RIGAUD's PhD (defended in 2014) [Report] & [Synopsis] & [Slides]
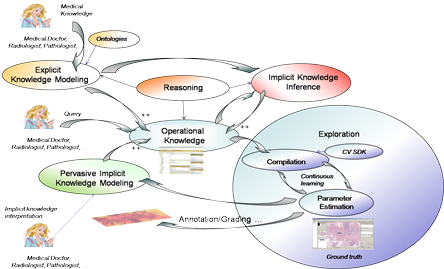
The explicit knowledge used by the pathologists need to be modeled and used in an integrated manner with multi-scale computer vision algorithms, in order to achieve a meaningful annotation in reasonable time.
Due to the huge size of the microscopic images (definition is about 60,000 x 50,000 pixels = 3000 megapixels — about 8 GB uncompressed for one virtual slide only at magnitude 40X having a resolution of 0.25 µm by pixel), and the time constraint (pathologists need about 5 to 10 minutes for a complete slide grading), this task is very challenging, necessitating fast multi-scale exploration algorithms, by considering a visual ontology dedicated to breast cancer grading and new continuous learning. approaches.
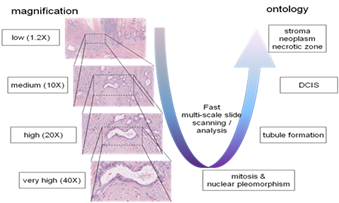
Fig.4: Semantic multi-scale image exploration
Integrating pathology and radiology information into the same PACS (Picture Archiving and Communication System) / EMR (Electronic Medical Record) platform.
Model (including physics, chemical and biological mathematical modeling and ontologies) allowing to interact/exchange between the micro- and the macro-modalities and associated physiologic knowledge.
The scientific impact and in terms of new opportunities for the future generation of PACS is considerable, bringing a new way of doing histopathology in the near future.
PhD work of Adina Tutac: PhD report & PhD oral defense slides
Develop a novel semiologic ontology modeling approach for breast cancer grading (BCG).
Transform the virtual microscope into a cognitive virtual microscope, by a synergy between medical knowledge management and computer vision.
Fig.5: Cognitive exploration of the images in histopathology
PhD work of Roxana Teodorescu: PhD report & PhD oral defense slides
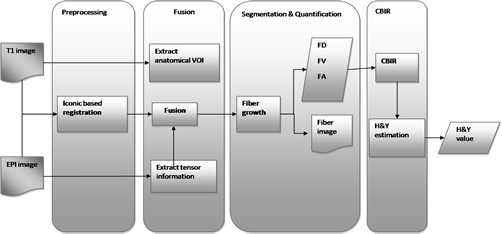
The purpose of our study is the prognosis of Parkinson’s disease by correlating H&Y tests results with specific brain features from mulitple MRI modalities. Using the DTI images, we take into account the advantages/ disadvantages offered by each of the modalities. From the T1 images, we extract the red nucleus area, containing the Substantia Negra (SN). This imaging type has a well defined anatomy and the quality of the image is better than the other DTIs. From the FA images we extract the Putamen (Put) area as well as the Globus Pallidus (GP) area. These areas represent the destination of the neural tracts that start from SN and belong to the neuro-motor fibers. These fibers are the first affected by the lack of dopamine at the brain level. The FA image offers a better delimitation for the Put and GP.
We use MRI multimodal image analysis for feature extraction and perform a fusion between these characteristics for classification purposes. Using the classified data, we compute a score meant to give us a variation in time of the disease that we use next for prognosis. First we find the differences between Parkinson’s disease (PD) affected patients and the control patients. A more refined step is related to the stage of the disease and to find the differences between these patients. The final step is represented by the global analysis of the features and the temporal evolution of the disease related to its severity.
Fig.6: PD prediction workflow
This system offers the perspective of classifying patients according to the PD severity using our image analysis system. This option comes to complete the cognitive tests applied as it is correlated with them.
|
Extracted Mid Brain |
Extracted Putamen |
Fibers and PD-relevant ROIs |
Fig. 7. Multimodal image indexing & analysis for early detection of Parkinson’s Disease - Parkinson's Disease Dedicated Atlas (ImageJ plugin)
Establish a link between the specific features of the Parkinson's Disease and the ground truth (H&Y) in order to extrapolate the results and establish a prognosis policy.